Support Vector Machines
Contents
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from sklearn.datasets import make_blobs, load_breast_cancer
from sklearn.model_selection import cross_val_score, train_test_split
from sklearn.svm import SVC
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import confusion_matrix, classification_report
Support Vector Machines#
A type of discriminate machine learning model
They try and build a plane in space that separates examples that belong to different classes with the widest possible margin
New examples that are mapped onto that space get “classified” based on which side of the boundary they fall on
Example: Random Data#
# creating datasets X containing n_samples
# Y containing two classes
X, Y = make_blobs(n_samples=500, centers=2,
random_state=0, cluster_std=0.40)
# plotting scatters
plt.scatter(X[:, 0], X[:, 1], c=Y, s=50, cmap='RdBu');
plt.show()
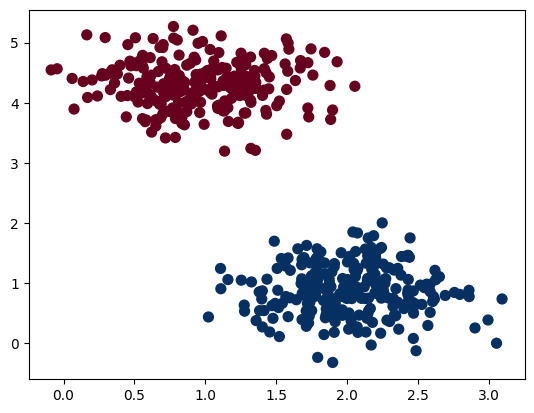
In this case, it is very easy to classify the points with a linear boundary
# creating line space between -1 to 3.5
xfit = np.linspace(-1, 3.5)
# plotting scatter
plt.scatter(X[:, 0], X[:, 1], c=Y, s=50, cmap='RdBu')
# plot a line between the different sets of data
for m, b, d in [(1, 0.65, 0.33), (0.5, 1.6, 0.55), (-0.2, 2.9, 0.2)]:
yfit = m * xfit + b
plt.plot(xfit, yfit, '-k')
plt.fill_between(xfit, yfit - d, yfit + d, edgecolor='none',
color='#AAAAAA', alpha=0.4)
plt.xlim(-1, 3.5);
plt.show()
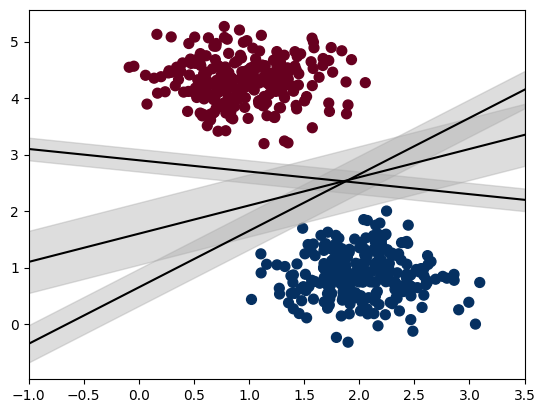
SVMs try and find the widest possible perpendicular distance between the dividing vector and the points
Example: Breast Cancer Data#
Step 1: Load the Data#
We’ll use the built-in breast cancer dataset from Scikit-Learn. We can get with the load function:
cancer = load_breast_cancer()
Step 2: Understanding the Data#
The data set is presented in a dictionary form:
cancer.keys()
dict_keys(['data', 'target', 'frame', 'target_names', 'DESCR', 'feature_names', 'filename', 'data_module'])
The DESCR contains a description of the information in the dataset
print(cancer['DESCR'])
.. _breast_cancer_dataset:
Breast cancer wisconsin (diagnostic) dataset
--------------------------------------------
**Data Set Characteristics:**
:Number of Instances: 569
:Number of Attributes: 30 numeric, predictive attributes and the class
:Attribute Information:
- radius (mean of distances from center to points on the perimeter)
- texture (standard deviation of gray-scale values)
- perimeter
- area
- smoothness (local variation in radius lengths)
- compactness (perimeter^2 / area - 1.0)
- concavity (severity of concave portions of the contour)
- concave points (number of concave portions of the contour)
- symmetry
- fractal dimension ("coastline approximation" - 1)
The mean, standard error, and "worst" or largest (mean of the three
worst/largest values) of these features were computed for each image,
resulting in 30 features. For instance, field 0 is Mean Radius, field
10 is Radius SE, field 20 is Worst Radius.
- class:
- WDBC-Malignant
- WDBC-Benign
:Summary Statistics:
===================================== ====== ======
Min Max
===================================== ====== ======
radius (mean): 6.981 28.11
texture (mean): 9.71 39.28
perimeter (mean): 43.79 188.5
area (mean): 143.5 2501.0
smoothness (mean): 0.053 0.163
compactness (mean): 0.019 0.345
concavity (mean): 0.0 0.427
concave points (mean): 0.0 0.201
symmetry (mean): 0.106 0.304
fractal dimension (mean): 0.05 0.097
radius (standard error): 0.112 2.873
texture (standard error): 0.36 4.885
perimeter (standard error): 0.757 21.98
area (standard error): 6.802 542.2
smoothness (standard error): 0.002 0.031
compactness (standard error): 0.002 0.135
concavity (standard error): 0.0 0.396
concave points (standard error): 0.0 0.053
symmetry (standard error): 0.008 0.079
fractal dimension (standard error): 0.001 0.03
radius (worst): 7.93 36.04
texture (worst): 12.02 49.54
perimeter (worst): 50.41 251.2
area (worst): 185.2 4254.0
smoothness (worst): 0.071 0.223
compactness (worst): 0.027 1.058
concavity (worst): 0.0 1.252
concave points (worst): 0.0 0.291
symmetry (worst): 0.156 0.664
fractal dimension (worst): 0.055 0.208
===================================== ====== ======
:Missing Attribute Values: None
:Class Distribution: 212 - Malignant, 357 - Benign
:Creator: Dr. William H. Wolberg, W. Nick Street, Olvi L. Mangasarian
:Donor: Nick Street
:Date: November, 1995
This is a copy of UCI ML Breast Cancer Wisconsin (Diagnostic) datasets.
https://goo.gl/U2Uwz2
Features are computed from a digitized image of a fine needle
aspirate (FNA) of a breast mass. They describe
characteristics of the cell nuclei present in the image.
Separating plane described above was obtained using
Multisurface Method-Tree (MSM-T) [K. P. Bennett, "Decision Tree
Construction Via Linear Programming." Proceedings of the 4th
Midwest Artificial Intelligence and Cognitive Science Society,
pp. 97-101, 1992], a classification method which uses linear
programming to construct a decision tree. Relevant features
were selected using an exhaustive search in the space of 1-4
features and 1-3 separating planes.
The actual linear program used to obtain the separating plane
in the 3-dimensional space is that described in:
[K. P. Bennett and O. L. Mangasarian: "Robust Linear
Programming Discrimination of Two Linearly Inseparable Sets",
Optimization Methods and Software 1, 1992, 23-34].
This database is also available through the UW CS ftp server:
ftp ftp.cs.wisc.edu
cd math-prog/cpo-dataset/machine-learn/WDBC/
.. topic:: References
- W.N. Street, W.H. Wolberg and O.L. Mangasarian. Nuclear feature extraction
for breast tumor diagnosis. IS&T/SPIE 1993 International Symposium on
Electronic Imaging: Science and Technology, volume 1905, pages 861-870,
San Jose, CA, 1993.
- O.L. Mangasarian, W.N. Street and W.H. Wolberg. Breast cancer diagnosis and
prognosis via linear programming. Operations Research, 43(4), pages 570-577,
July-August 1995.
- W.H. Wolberg, W.N. Street, and O.L. Mangasarian. Machine learning techniques
to diagnose breast cancer from fine-needle aspirates. Cancer Letters 77 (1994)
163-171.
# A list of the feature names
cancer['feature_names']
array(['mean radius', 'mean texture', 'mean perimeter', 'mean area',
'mean smoothness', 'mean compactness', 'mean concavity',
'mean concave points', 'mean symmetry', 'mean fractal dimension',
'radius error', 'texture error', 'perimeter error', 'area error',
'smoothness error', 'compactness error', 'concavity error',
'concave points error', 'symmetry error',
'fractal dimension error', 'worst radius', 'worst texture',
'worst perimeter', 'worst area', 'worst smoothness',
'worst compactness', 'worst concavity', 'worst concave points',
'worst symmetry', 'worst fractal dimension'], dtype='<U23')
Step 3: Building the Data Structure for Training#
df_feat = pd.DataFrame(cancer['data'],columns=cancer['feature_names'])
df_feat.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 569 entries, 0 to 568
Data columns (total 30 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 mean radius 569 non-null float64
1 mean texture 569 non-null float64
2 mean perimeter 569 non-null float64
3 mean area 569 non-null float64
4 mean smoothness 569 non-null float64
5 mean compactness 569 non-null float64
6 mean concavity 569 non-null float64
7 mean concave points 569 non-null float64
8 mean symmetry 569 non-null float64
9 mean fractal dimension 569 non-null float64
10 radius error 569 non-null float64
11 texture error 569 non-null float64
12 perimeter error 569 non-null float64
13 area error 569 non-null float64
14 smoothness error 569 non-null float64
15 compactness error 569 non-null float64
16 concavity error 569 non-null float64
17 concave points error 569 non-null float64
18 symmetry error 569 non-null float64
19 fractal dimension error 569 non-null float64
20 worst radius 569 non-null float64
21 worst texture 569 non-null float64
22 worst perimeter 569 non-null float64
23 worst area 569 non-null float64
24 worst smoothness 569 non-null float64
25 worst compactness 569 non-null float64
26 worst concavity 569 non-null float64
27 worst concave points 569 non-null float64
28 worst symmetry 569 non-null float64
29 worst fractal dimension 569 non-null float64
dtypes: float64(30)
memory usage: 133.5 KB
# Views how the labels are structured
cancer['target']
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0,
0, 0, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0,
1, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 0, 0,
1, 1, 1, 0, 1, 1, 0, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 1, 0, 1,
1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 1, 0,
0, 1, 0, 0, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1,
1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 0, 0, 1, 1, 0, 0, 1, 1, 1,
1, 0, 1, 1, 0, 0, 0, 1, 0, 1, 0, 1, 1, 1, 0, 1, 1, 0, 0, 1, 0, 0,
0, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 1, 0, 0, 0, 0, 1, 1, 0, 0,
1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 1, 1,
1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 1, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 0, 1, 1, 0, 1, 0, 0, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0,
0, 1, 1, 1, 1, 0, 1, 0, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0,
0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0,
1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 0, 0, 1, 1,
1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 0,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 1, 1, 1,
1, 0, 1, 1, 0, 1, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0,
1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1,
1, 1, 1, 0, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 1, 0, 1, 1,
1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1])
df_target = pd.DataFrame(cancer['target'],columns=['Cancer'])
Step 4: Validate the Data Structure#
df_feat.head()
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst radius | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 17.99 | 10.38 | 122.80 | 1001.0 | 0.11840 | 0.27760 | 0.3001 | 0.14710 | 0.2419 | 0.07871 | ... | 25.38 | 17.33 | 184.60 | 2019.0 | 0.1622 | 0.6656 | 0.7119 | 0.2654 | 0.4601 | 0.11890 |
| 1 | 20.57 | 17.77 | 132.90 | 1326.0 | 0.08474 | 0.07864 | 0.0869 | 0.07017 | 0.1812 | 0.05667 | ... | 24.99 | 23.41 | 158.80 | 1956.0 | 0.1238 | 0.1866 | 0.2416 | 0.1860 | 0.2750 | 0.08902 |
| 2 | 19.69 | 21.25 | 130.00 | 1203.0 | 0.10960 | 0.15990 | 0.1974 | 0.12790 | 0.2069 | 0.05999 | ... | 23.57 | 25.53 | 152.50 | 1709.0 | 0.1444 | 0.4245 | 0.4504 | 0.2430 | 0.3613 | 0.08758 |
| 3 | 11.42 | 20.38 | 77.58 | 386.1 | 0.14250 | 0.28390 | 0.2414 | 0.10520 | 0.2597 | 0.09744 | ... | 14.91 | 26.50 | 98.87 | 567.7 | 0.2098 | 0.8663 | 0.6869 | 0.2575 | 0.6638 | 0.17300 |
| 4 | 20.29 | 14.34 | 135.10 | 1297.0 | 0.10030 | 0.13280 | 0.1980 | 0.10430 | 0.1809 | 0.05883 | ... | 22.54 | 16.67 | 152.20 | 1575.0 | 0.1374 | 0.2050 | 0.4000 | 0.1625 | 0.2364 | 0.07678 |
5 rows × 30 columns
Generally, at this stage, one would visualize that data using domain knowledge
Step 5: Test-Train Split#
X_train, X_test, y_train, y_test = train_test_split(df_feat, np.ravel(df_target), test_size=0.30, random_state=101)
Step 6: Train the Model#
model = SVC(gamma='auto')
model.fit(X_train,y_train)
SVC(gamma='auto')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
SVC(gamma='auto')
X_train
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst radius | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 178 | 13.010 | 22.22 | 82.01 | 526.4 | 0.06251 | 0.01938 | 0.001595 | 0.001852 | 0.1395 | 0.05234 | ... | 14.00 | 29.02 | 88.18 | 608.8 | 0.08125 | 0.03432 | 0.007977 | 0.009259 | 0.2295 | 0.05843 |
| 421 | 14.690 | 13.98 | 98.22 | 656.1 | 0.10310 | 0.18360 | 0.145000 | 0.063000 | 0.2086 | 0.07406 | ... | 16.46 | 18.34 | 114.10 | 809.2 | 0.13120 | 0.36350 | 0.321900 | 0.110800 | 0.2827 | 0.09208 |
| 57 | 14.710 | 21.59 | 95.55 | 656.9 | 0.11370 | 0.13650 | 0.129300 | 0.081230 | 0.2027 | 0.06758 | ... | 17.87 | 30.70 | 115.70 | 985.5 | 0.13680 | 0.42900 | 0.358700 | 0.183400 | 0.3698 | 0.10940 |
| 514 | 15.050 | 19.07 | 97.26 | 701.9 | 0.09215 | 0.08597 | 0.074860 | 0.043350 | 0.1561 | 0.05915 | ... | 17.58 | 28.06 | 113.80 | 967.0 | 0.12460 | 0.21010 | 0.286600 | 0.112000 | 0.2282 | 0.06954 |
| 548 | 9.683 | 19.34 | 61.05 | 285.7 | 0.08491 | 0.05030 | 0.023370 | 0.009615 | 0.1580 | 0.06235 | ... | 10.93 | 25.59 | 69.10 | 364.2 | 0.11990 | 0.09546 | 0.093500 | 0.038460 | 0.2552 | 0.07920 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 552 | 12.770 | 29.43 | 81.35 | 507.9 | 0.08276 | 0.04234 | 0.019970 | 0.014990 | 0.1539 | 0.05637 | ... | 13.87 | 36.00 | 88.10 | 594.7 | 0.12340 | 0.10640 | 0.086530 | 0.064980 | 0.2407 | 0.06484 |
| 393 | 21.610 | 22.28 | 144.40 | 1407.0 | 0.11670 | 0.20870 | 0.281000 | 0.156200 | 0.2162 | 0.06606 | ... | 26.23 | 28.74 | 172.00 | 2081.0 | 0.15020 | 0.57170 | 0.705300 | 0.242200 | 0.3828 | 0.10070 |
| 75 | 16.070 | 19.65 | 104.10 | 817.7 | 0.09168 | 0.08424 | 0.097690 | 0.066380 | 0.1798 | 0.05391 | ... | 19.77 | 24.56 | 128.80 | 1223.0 | 0.15000 | 0.20450 | 0.282900 | 0.152000 | 0.2650 | 0.06387 |
| 337 | 18.770 | 21.43 | 122.90 | 1092.0 | 0.09116 | 0.14020 | 0.106000 | 0.060900 | 0.1953 | 0.06083 | ... | 24.54 | 34.37 | 161.10 | 1873.0 | 0.14980 | 0.48270 | 0.463400 | 0.204800 | 0.3679 | 0.09870 |
| 523 | 13.710 | 18.68 | 88.73 | 571.0 | 0.09916 | 0.10700 | 0.053850 | 0.037830 | 0.1714 | 0.06843 | ... | 15.11 | 25.63 | 99.43 | 701.9 | 0.14250 | 0.25660 | 0.193500 | 0.128400 | 0.2849 | 0.09031 |
398 rows × 30 columns
Step 7: Visualize the Results#
predictions = model.predict(X_test)
cm = confusion_matrix(y_test,predictions)
plt.clf()
plt.imshow(cm, interpolation='nearest', cmap=plt.cm.viridis)
classNames = ['Negative','Positive']
plt.title('Cancer Confusion Matrix')
plt.ylabel('True label')
plt.xlabel('Predicted label')
tick_marks = np.arange(len(classNames))
plt.xticks(tick_marks, classNames, rotation=45)
plt.yticks(tick_marks, classNames)
s = [['TN','FP'], ['FN', 'TP']]
for i in range(2):
for j in range(2):
plt.text(j,i, str(s[i][j])+" = "+str(cm[i][j]))
plt.show()
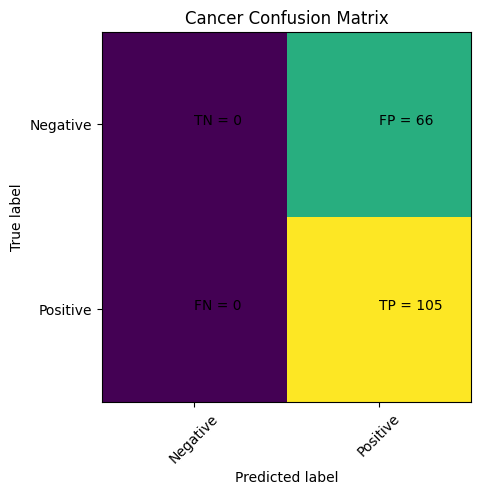
print(classification_report(y_test,predictions))
precision recall f1-score support
0 0.00 0.00 0.00 66
1 0.61 1.00 0.76 105
accuracy 0.61 171
macro avg 0.31 0.50 0.38 171
weighted avg 0.38 0.61 0.47 171
C:\Users\jca92\.conda\envs\jupyterbook\lib\site-packages\sklearn\metrics\_classification.py:1334: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, msg_start, len(result))
C:\Users\jca92\.conda\envs\jupyterbook\lib\site-packages\sklearn\metrics\_classification.py:1334: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, msg_start, len(result))
C:\Users\jca92\.conda\envs\jupyterbook\lib\site-packages\sklearn\metrics\_classification.py:1334: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, msg_start, len(result))
This is not good … Our Machine Thinks Everyone has Cancer!!!!
What did we do wrong?
Step 4a: Scaling the Data#
When models fail:
Check how the data was normalized
Optimize the model parameters
df_feat.head()
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst radius | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 17.99 | 10.38 | 122.80 | 1001.0 | 0.11840 | 0.27760 | 0.3001 | 0.14710 | 0.2419 | 0.07871 | ... | 25.38 | 17.33 | 184.60 | 2019.0 | 0.1622 | 0.6656 | 0.7119 | 0.2654 | 0.4601 | 0.11890 |
| 1 | 20.57 | 17.77 | 132.90 | 1326.0 | 0.08474 | 0.07864 | 0.0869 | 0.07017 | 0.1812 | 0.05667 | ... | 24.99 | 23.41 | 158.80 | 1956.0 | 0.1238 | 0.1866 | 0.2416 | 0.1860 | 0.2750 | 0.08902 |
| 2 | 19.69 | 21.25 | 130.00 | 1203.0 | 0.10960 | 0.15990 | 0.1974 | 0.12790 | 0.2069 | 0.05999 | ... | 23.57 | 25.53 | 152.50 | 1709.0 | 0.1444 | 0.4245 | 0.4504 | 0.2430 | 0.3613 | 0.08758 |
| 3 | 11.42 | 20.38 | 77.58 | 386.1 | 0.14250 | 0.28390 | 0.2414 | 0.10520 | 0.2597 | 0.09744 | ... | 14.91 | 26.50 | 98.87 | 567.7 | 0.2098 | 0.8663 | 0.6869 | 0.2575 | 0.6638 | 0.17300 |
| 4 | 20.29 | 14.34 | 135.10 | 1297.0 | 0.10030 | 0.13280 | 0.1980 | 0.10430 | 0.1809 | 0.05883 | ... | 22.54 | 16.67 | 152.20 | 1575.0 | 0.1374 | 0.2050 | 0.4000 | 0.1625 | 0.2364 | 0.07678 |
5 rows × 30 columns
The features have vastly different scales which makes them weighted unequally
Standard Scaler#
Removes the mean and scales to a unit variance
\(\mu\) is the mean
\(\sigma\) is the standard deviation
The result is all features will have a mean of 0 and a variance of 1
# It is usually a good idea to scale the data for SVM training.
# We are cheating a bit in this example in scaling all of the data,
# instead of fitting the transformation on the training set and
# just applying it on the test set.
scaler = StandardScaler()
df_feat = scaler.fit_transform(df_feat)
df_feat = pd.DataFrame(df_feat,columns=cancer['feature_names'])
df_feat.head()
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst radius | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.097064 | -2.073335 | 1.269934 | 0.984375 | 1.568466 | 3.283515 | 2.652874 | 2.532475 | 2.217515 | 2.255747 | ... | 1.886690 | -1.359293 | 2.303601 | 2.001237 | 1.307686 | 2.616665 | 2.109526 | 2.296076 | 2.750622 | 1.937015 |
| 1 | 1.829821 | -0.353632 | 1.685955 | 1.908708 | -0.826962 | -0.487072 | -0.023846 | 0.548144 | 0.001392 | -0.868652 | ... | 1.805927 | -0.369203 | 1.535126 | 1.890489 | -0.375612 | -0.430444 | -0.146749 | 1.087084 | -0.243890 | 0.281190 |
| 2 | 1.579888 | 0.456187 | 1.566503 | 1.558884 | 0.942210 | 1.052926 | 1.363478 | 2.037231 | 0.939685 | -0.398008 | ... | 1.511870 | -0.023974 | 1.347475 | 1.456285 | 0.527407 | 1.082932 | 0.854974 | 1.955000 | 1.152255 | 0.201391 |
| 3 | -0.768909 | 0.253732 | -0.592687 | -0.764464 | 3.283553 | 3.402909 | 1.915897 | 1.451707 | 2.867383 | 4.910919 | ... | -0.281464 | 0.133984 | -0.249939 | -0.550021 | 3.394275 | 3.893397 | 1.989588 | 2.175786 | 6.046041 | 4.935010 |
| 4 | 1.750297 | -1.151816 | 1.776573 | 1.826229 | 0.280372 | 0.539340 | 1.371011 | 1.428493 | -0.009560 | -0.562450 | ... | 1.298575 | -1.466770 | 1.338539 | 1.220724 | 0.220556 | -0.313395 | 0.613179 | 0.729259 | -0.868353 | -0.397100 |
5 rows × 30 columns
Step 5: Test-Train Split#
X_train, X_test, y_train, y_test = train_test_split(df_feat, np.ravel(df_target), test_size=0.30, random_state=101)
Step 6: Train the Model#
model = SVC(gamma='auto')
model.fit(X_train,y_train)
SVC(gamma='auto')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
SVC(gamma='auto')
Step 7: Visualize the Results#
predictions = model.predict(X_test)
cm = confusion_matrix(y_test,predictions)
plt.clf()
plt.imshow(cm, interpolation='nearest', cmap=plt.cm.viridis)
classNames = ['Negative','Positive']
plt.title('Cancer Confusion Matrix')
plt.ylabel('True label')
plt.xlabel('Predicted label')
tick_marks = np.arange(len(classNames))
plt.xticks(tick_marks, classNames, rotation=45)
plt.yticks(tick_marks, classNames)
s = [['TN','FP'], ['FN', 'TP']]
for i in range(2):
for j in range(2):
plt.text(j,i, str(s[i][j])+" = "+str(cm[i][j]))
plt.show()
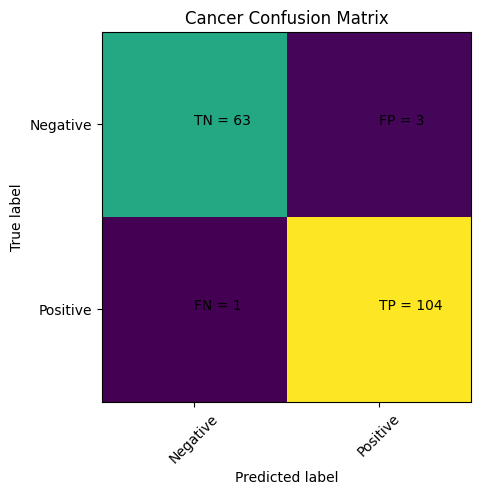
That looks way better



